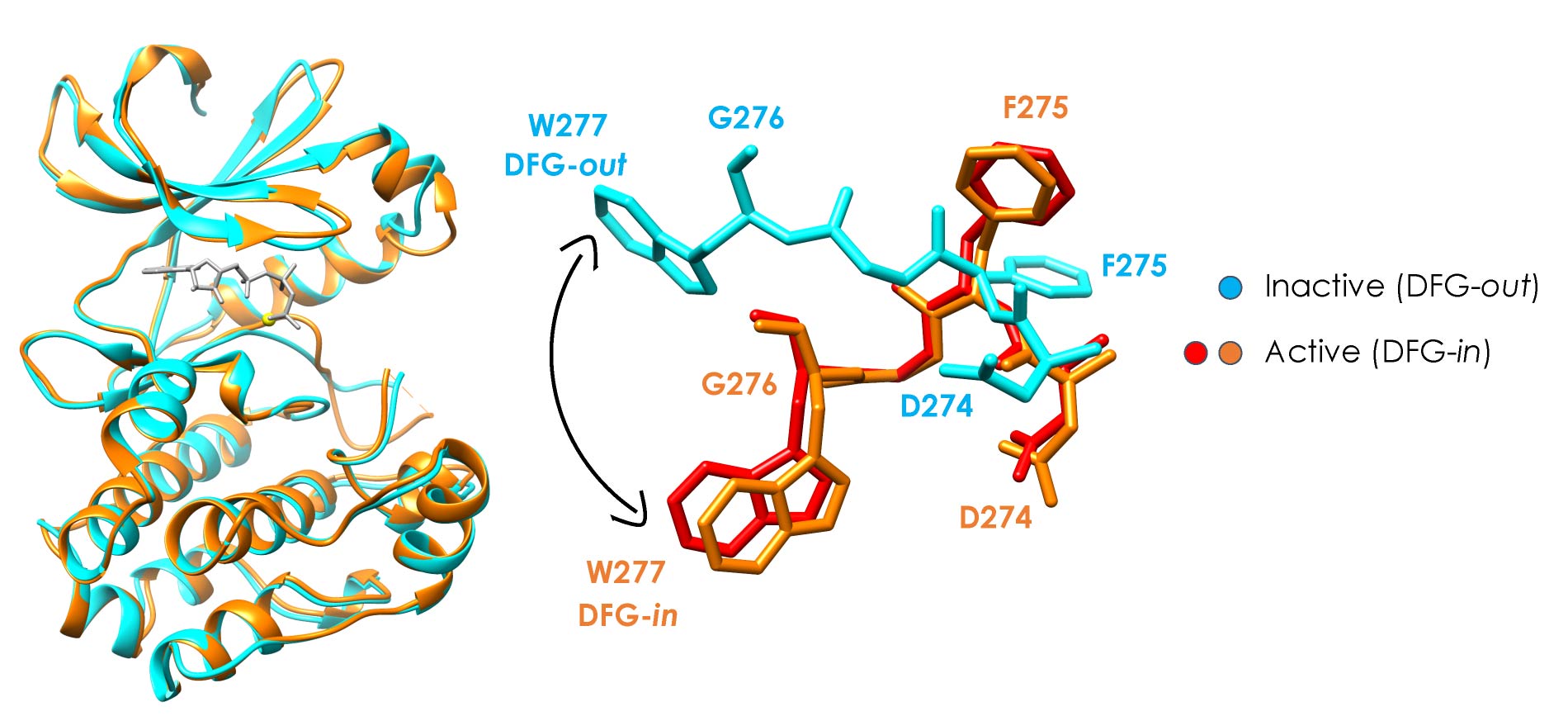
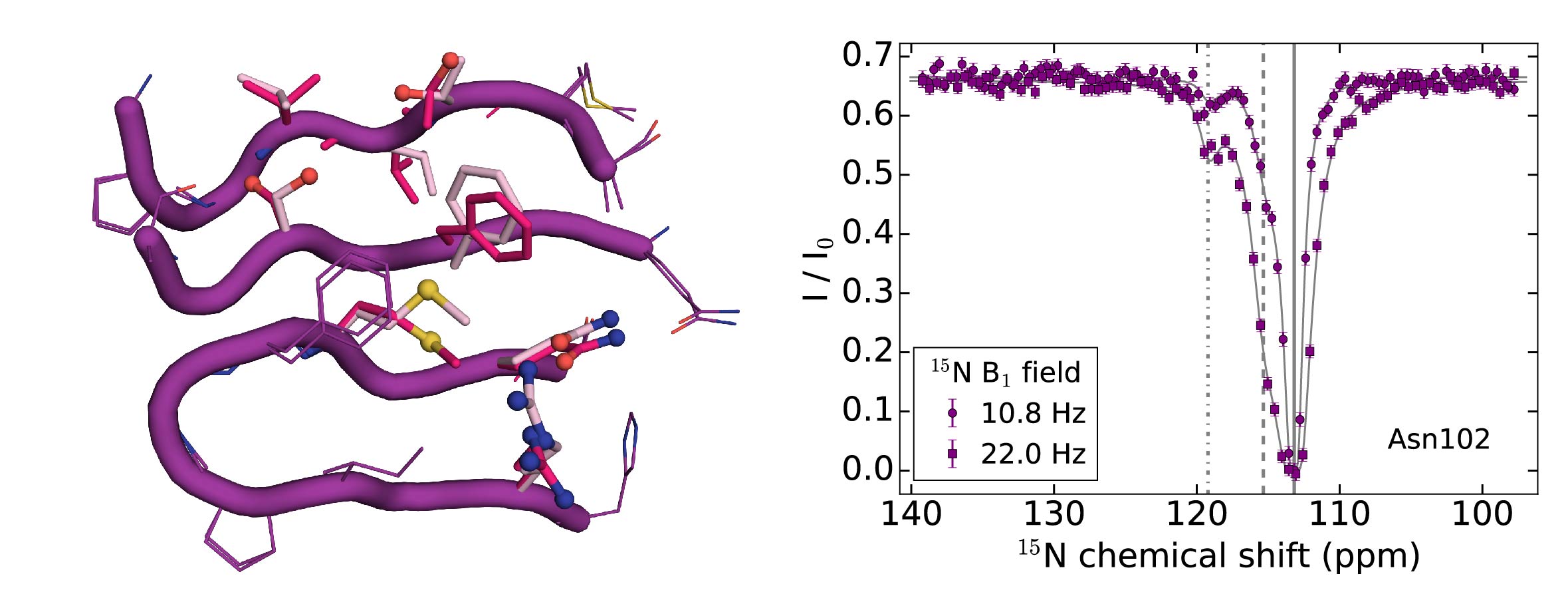
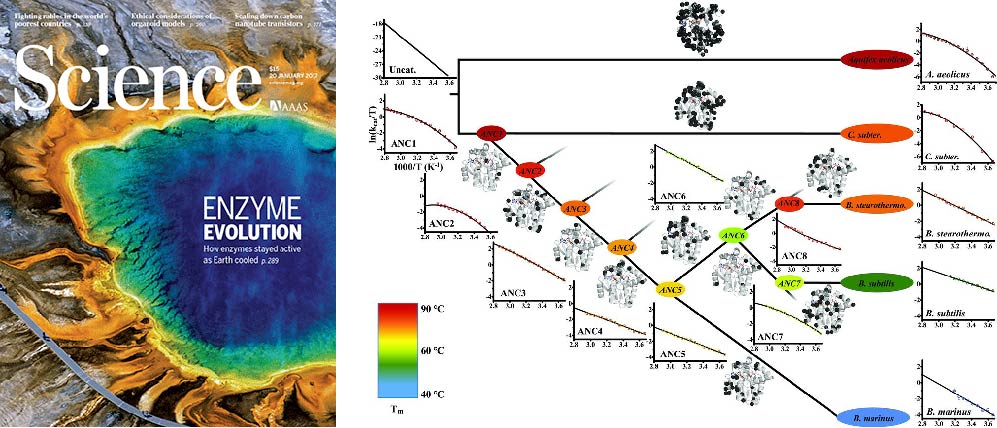
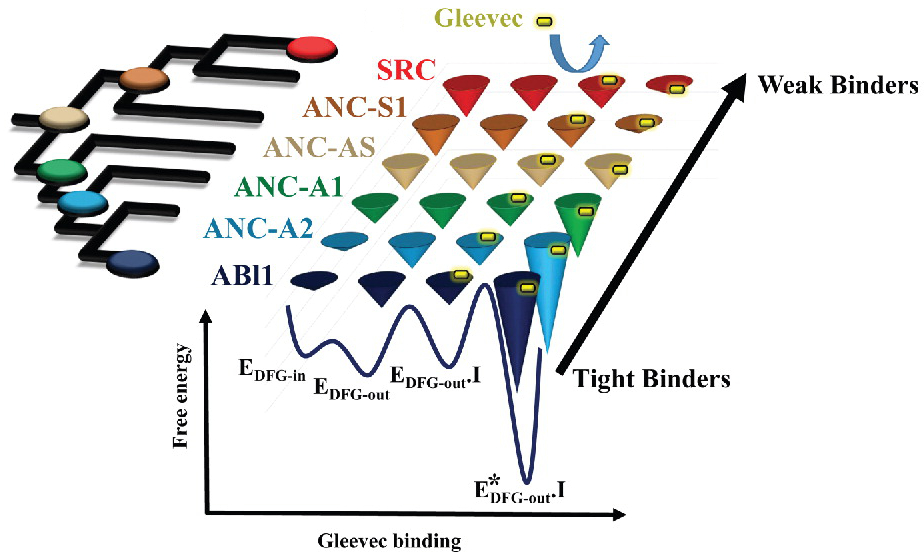
Publications
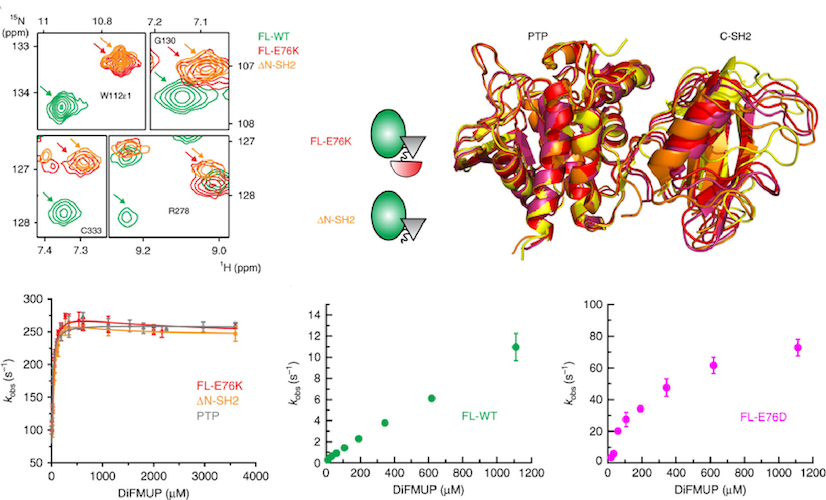
Glaser A, Pádua RAP, Ojoawo A, Sullivan C, Kern D. Phosphatase SHP2 pathogenic mutations enhance activity by altering conformational sampling bioRxiv 2025
- PMID:
Wayment-Steele HK#, El Nesr G#, Hettiarachchi R, Kariyawasam H, Ovchinnikov S, & Kern D. Learning millisecond protein dynamics from what is missing in NMR spectra. bioRxiv 2025
- PMID:
Wayment-Steele HK, Ovchinnikov S, Colwell L, Kern D. Does sequence clustering confound AlphaFold2? J Mol Biol 2025
- PMID: 40780395
Jara GE, Pontiggia F, Otten R, Agafonov RV, Marti MA & Kern D. Wide Trasition-State Ensemble as Key Component for Enzyme Catalysis. eLife 2025
- PMID: 39963964
- PMCID: PMC11835391
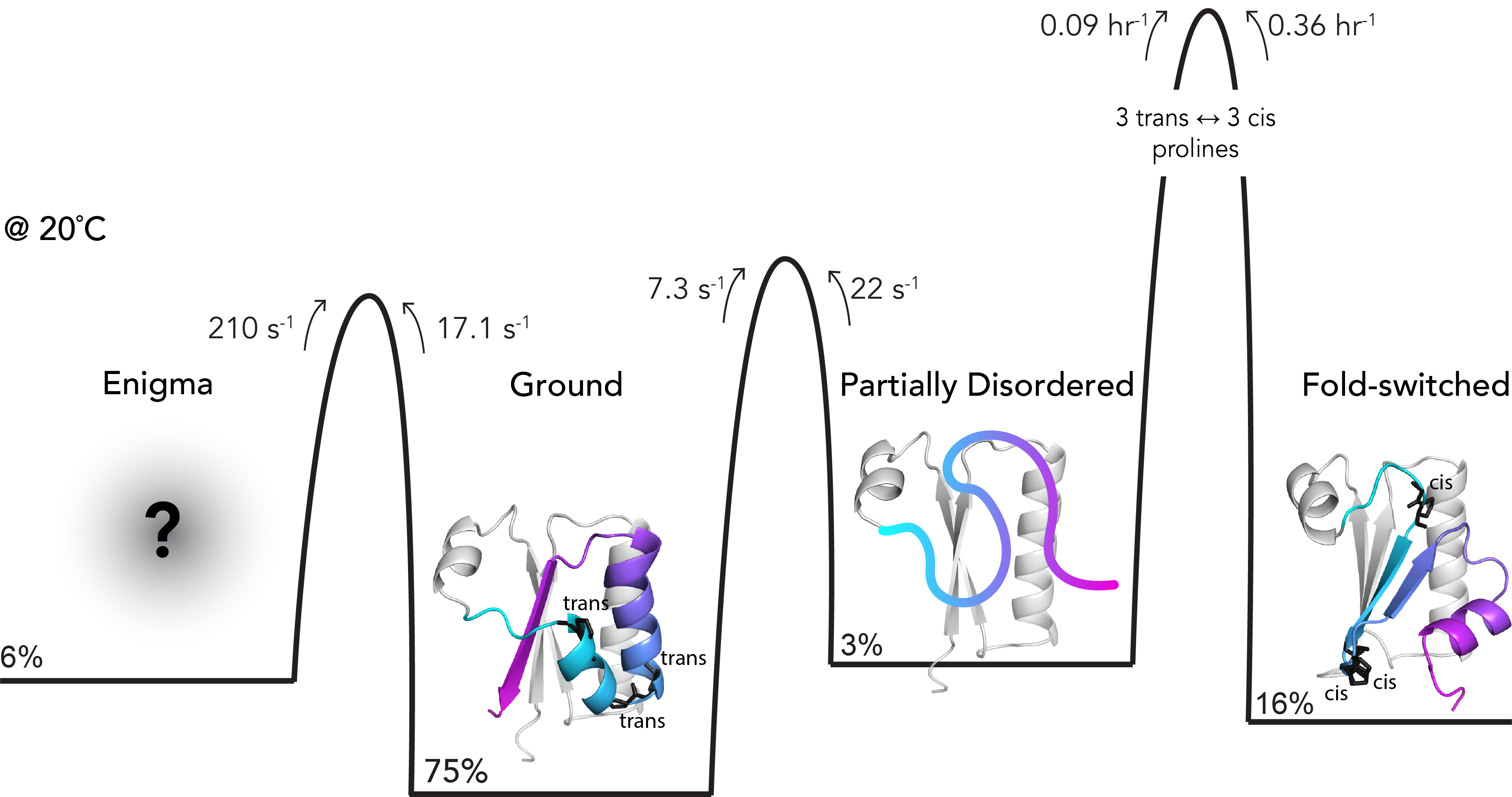
Wayment-Steele HK#, Otten R#, Pitsawong W#, Ojoawo A#, Glaser A, Calderone LA & Kern D. The conformational landscape of fold-switcher KaiB is tuned to the circadian rhythm timescale. Proc. Natl. Acad. Sci. US 2024
- PMID: 39475637
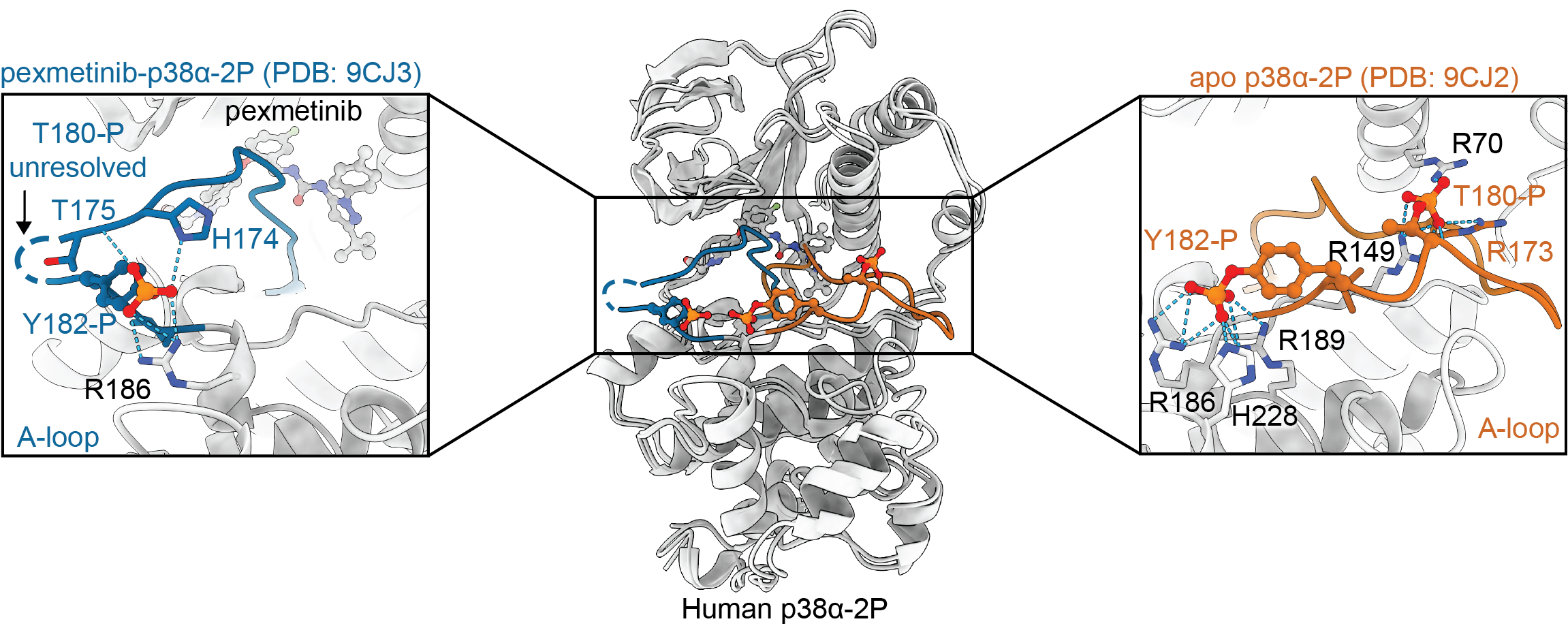
Stadnicki EJ, Ludewig H, Ramasamy PK, Wang X, Qiao Y, Kern D & Bradshaw N. Dual-Action Kinase Inhibitors Control p38α MAP Kinase Threonine Dephosphorylation. Proc. Natl. Acad. Sci. US 2024
- PMID:
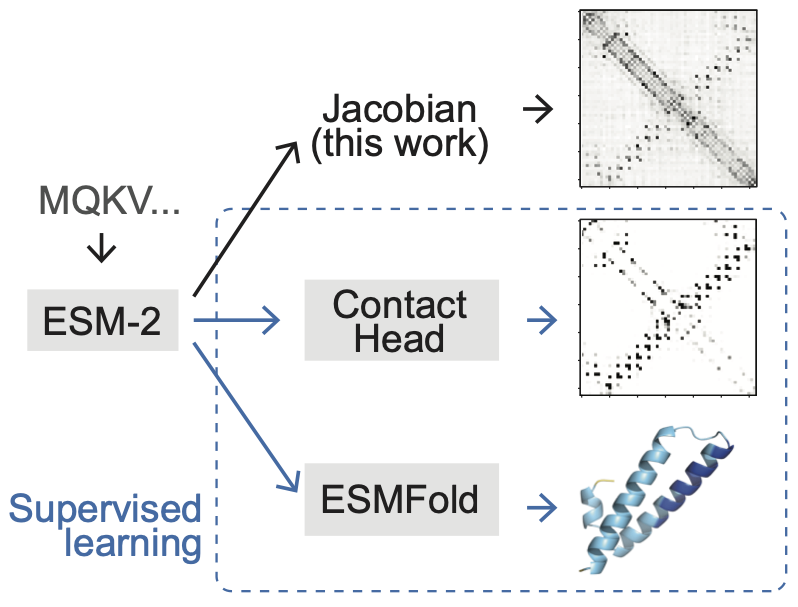
Zhang Z, Wayment-Steele HK, Brixi G, Wang H, Dal Peraro M, Kern D & Ovchinnikov S. Protein language models learn evolutionary statistics of interacting sequence motifs. Proc. Natl. Acad. Sci. US 2024
- PMID:
Wayment-Steele HK#, Ojoawo A#, Otten R, Apitz JM, Pitsawong W, Hoemberger M, Ovchinnikov S, Colwell L & Kern D. Predicting multiple conformations via sequence clustering and AlphaFold2. Nature 2023
- PMID: 37956700
- PMCID: PMC10808063
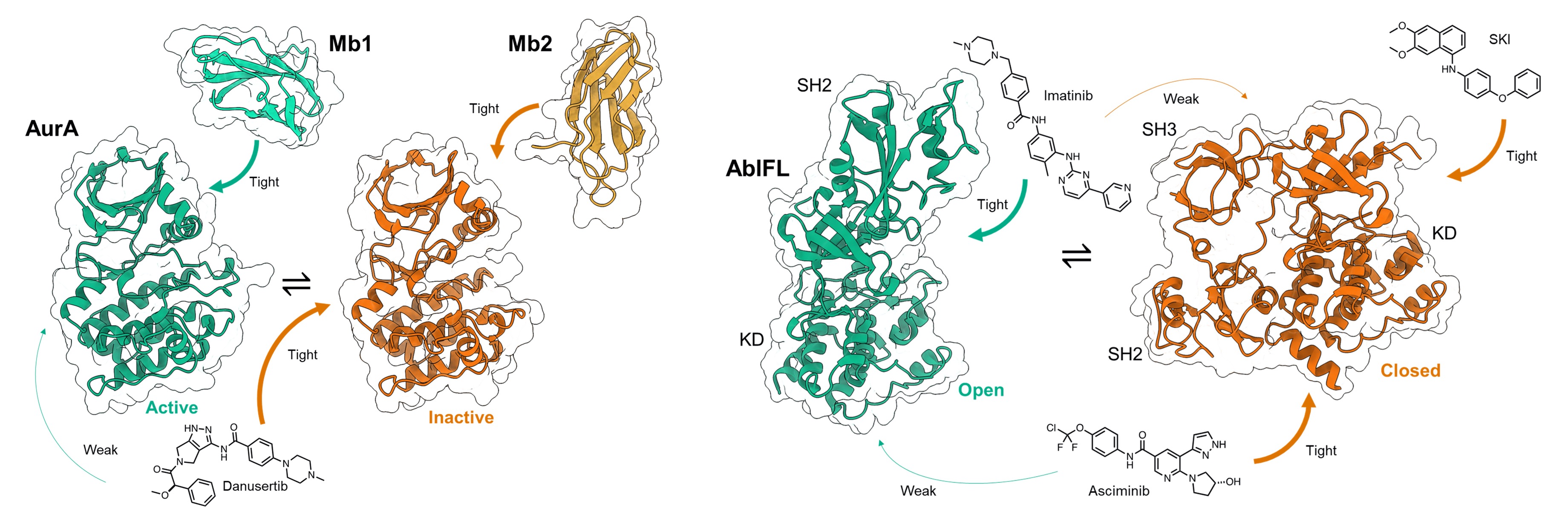
Kim C, Ludewig H, Hadzipasic A, Kutter S, Nguyen V & Kern D. A biophysical framework for double-drugging kinases. Proc. Natl. Acad. Sci. US 2023
- PMID: 37590418
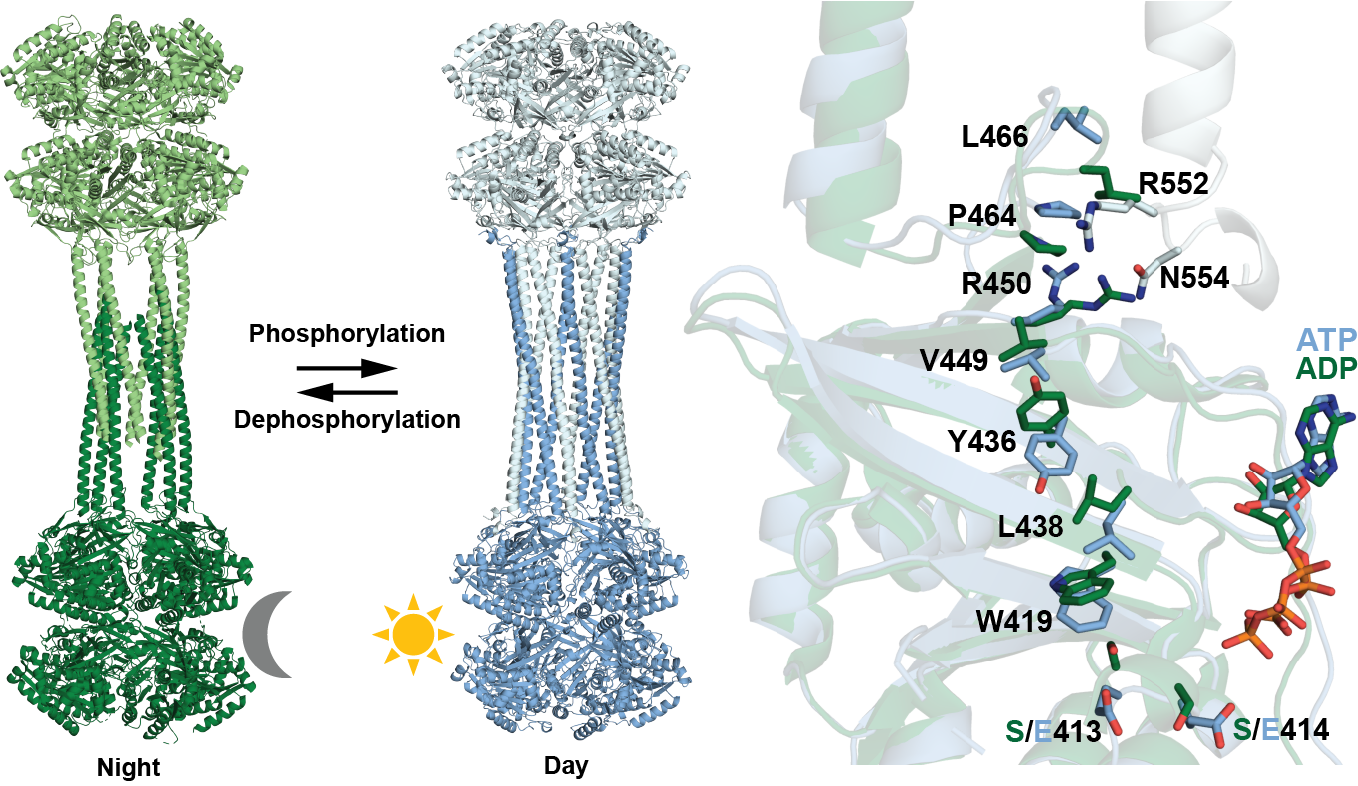
Pitsawong W, Pádua RAP, Grant T, Hoemberger M, Otten R, Bradshaw N, Grigorieff N & Kern D. From primordial clocks to circadian oscillators. Nature 2023
- PMID: 36949197
Stiller JB, Otten R, Häussinger D, Rieder PS, Theobald DL & Kern D. Structure determination of high-energy states in a dynamic protein ensemble. Nature 2022
- PMID: 35236984
- PMCID: Epub Ahead of Print
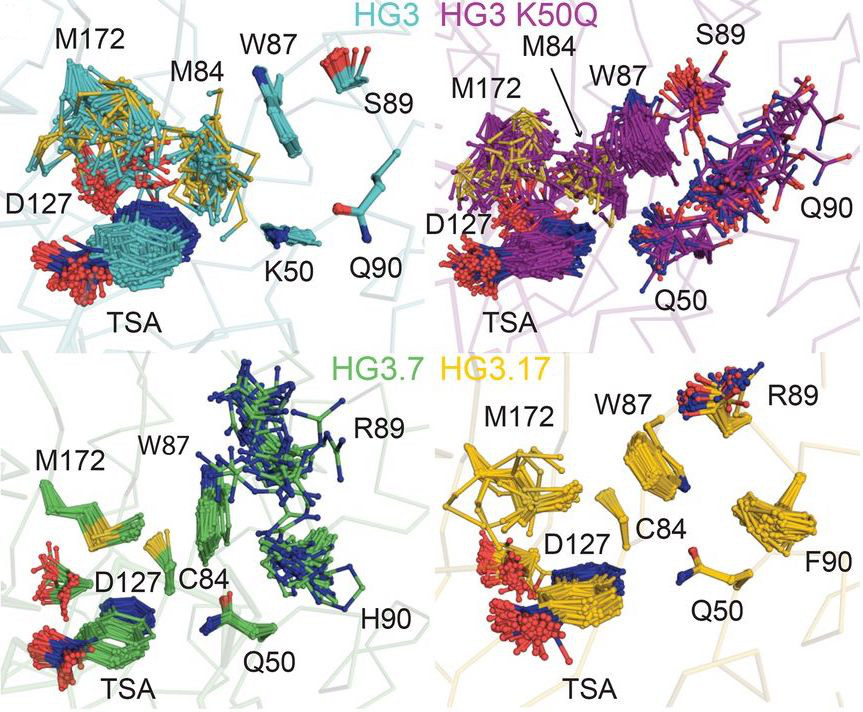
Otten R#, Pádua RAP#, Bunzel HA#, Nguyen V, Pitsawong W, Patterson M, Sui S, Perry SL, Cohen AE, Hilvert D & Kern D. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 2020
Hoemberger M, Pitsawong W & Kern D. Cumulative mechanism of several major imatinib-resistant mutations in Abl kinase. PNAS 2020
- PMID: 32719139
- PMCID: PMC7431045
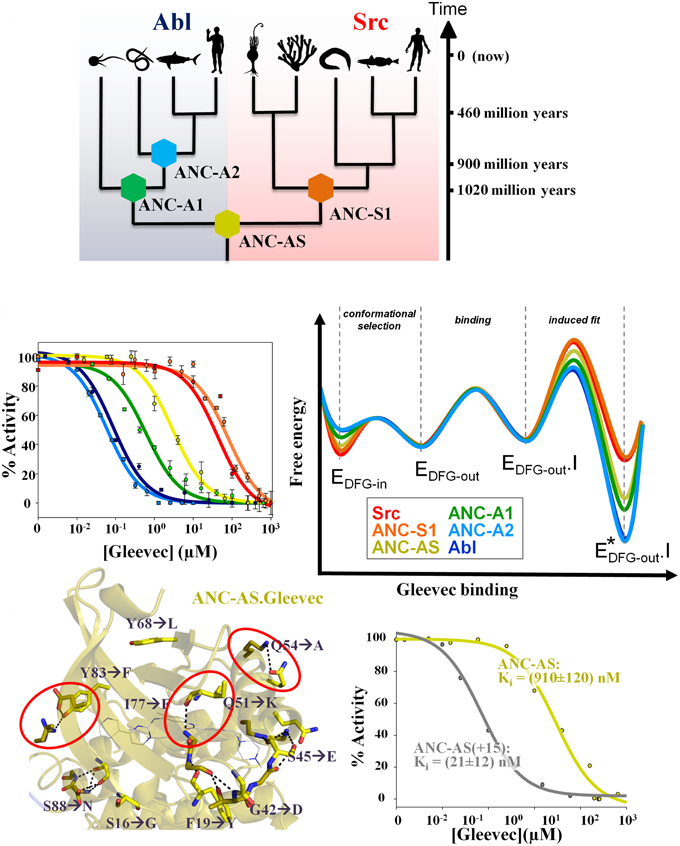
Hadzipasic A, Wilson C, Nguyen V, Kern N, Kim C, Pitsawong W, Villali J, Zheng Y & Kern D. Ancient origins of allosteric activation in a Ser-Thr kinase. Science 2020
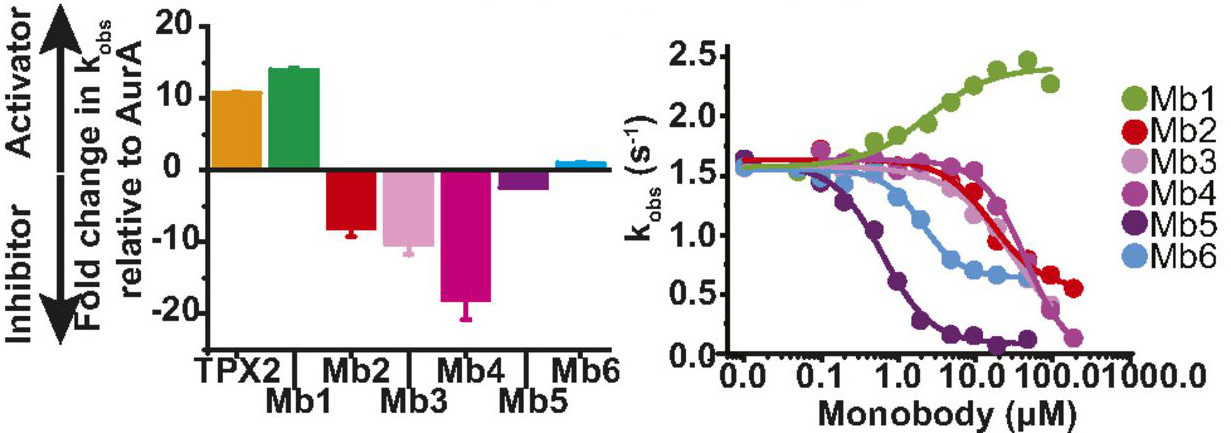
Zorba A#, Nguyen V#, Koide A, Hoemberger M, Zheng Y, Kutter S, Kim C, Koide S & Kern D. Allosteric Modulation of a Human Protein Kinase with Monobodies. Proc. Natl. Acad. Sci. USA 2019
- PMID: 31239342
- PMCID: PMC6628680
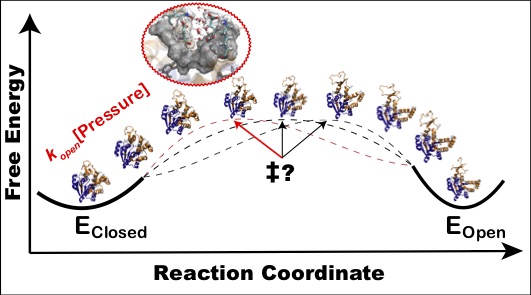
Stiller JB#, Kerns SJ#, Hoemberger M, Cho Y-J, Otten R, Hagan MF & Kern D. Probing the Transition State in Enzyme Catalysis by High-Pressure NMR Dynamics. Nat. Catal. 2019
Pádua RAP#, Sun Y#, Marko I, Pitsawong W, Stiller JB, Otten R & Kern D. Mechanism of Activating Mutations and Allosteric Drug Inhibition of the Phosphatase SHP2. Nat. Commun. 2018
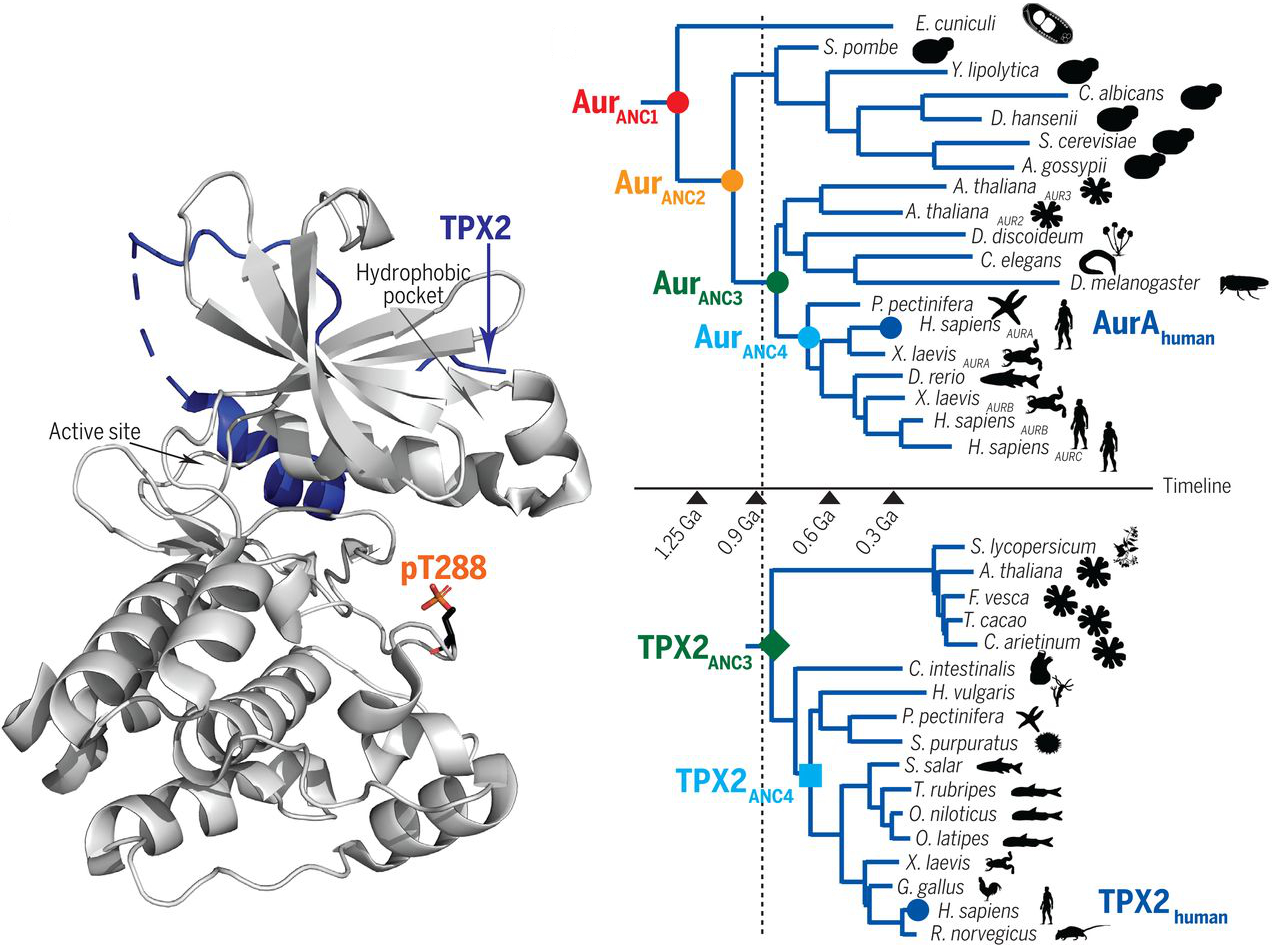
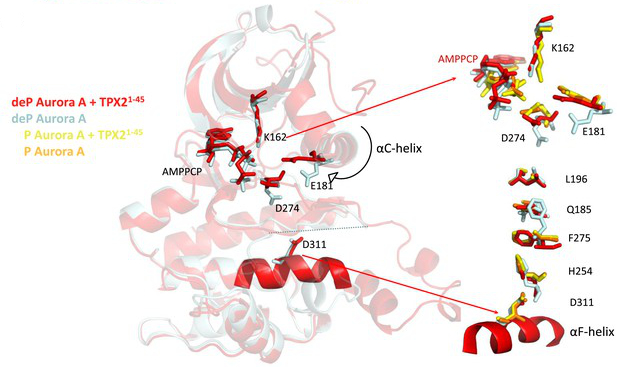
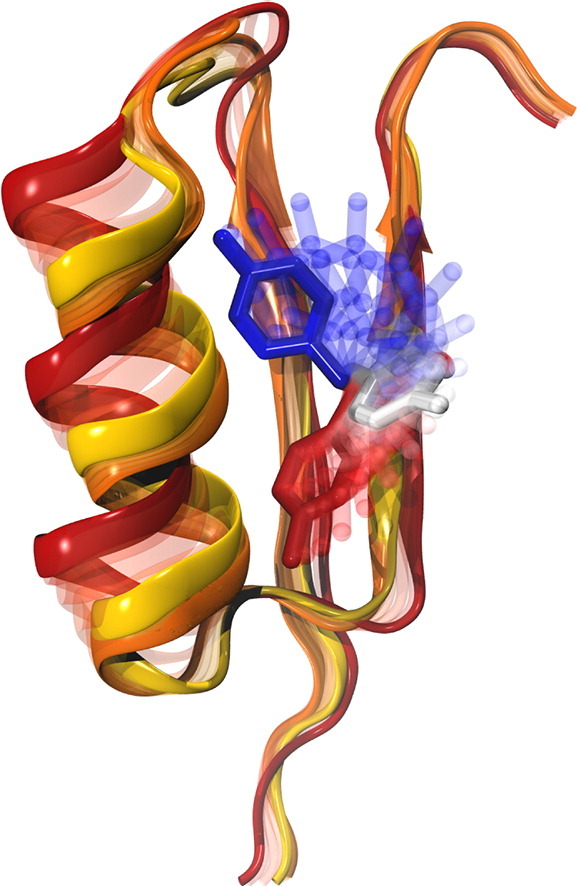
Pitsawong W#, Buosi V#, Otten R#, Agafonov RV#, Zorba A, Kern N, Kutter S, Kern G, Pádua RAP, Meniche X & Kern D. Dynamics of Human Protein Kinase Aurora A Linked to Drug Selectivity. eLife 2018
Otten R#, Liu L#, Kenner LR, Clarkson MW, Mavor D, Tawfik DS, Kern D & Fraser JS. Rescue of Conformational Dynamics in Enzyme Catalysis by Directed Evolution. Nat. Commun. 2018
Nguyen V#, Wilson C#, Hoemberger M, Stiller JB, Agafonov RV, Kutter S, English J, Theobald D & Kern D. Evolutionary Drivers of Thermoadaptation in Enzyme Catalysis. Science 2017
Chakrabarti KS#, Agafonov RV#, Pontiggia F, Otten R, Higgins MK, Schertler GF, Oprian DD & Kern D. Conformational Selection in a Protein-Protein Interaction Revealed by Dynamic Pathway Analysis. Cell Reports 2016
- PMID: 26725117
- PMCID: PMC4706811
Wilson C, Agafonov RV & Kern D. Drug targets evolve, and so should the methods. Mol. Cell Oncol. 2016
- PMID: 27652309
- PMCID: PMC4972104
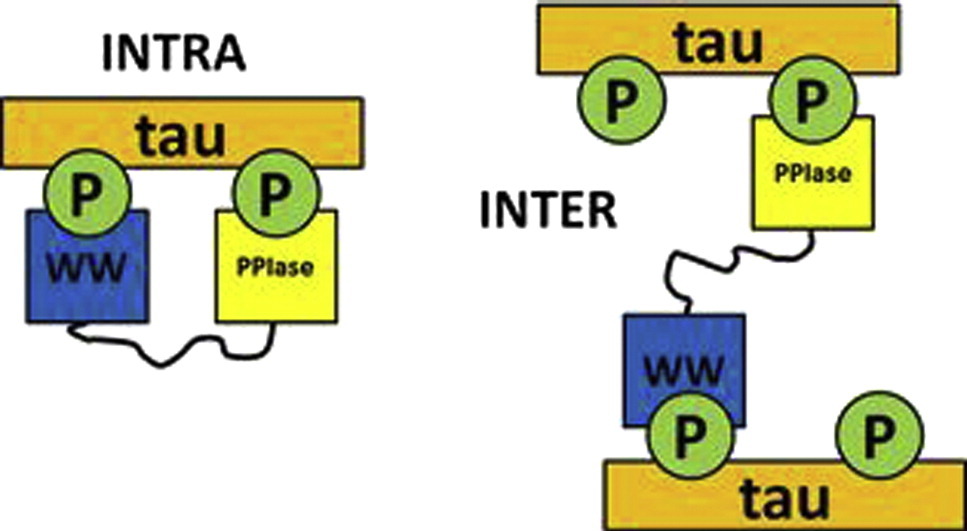
Eichner T, Kutter S, Labeikovsky W, Buosi V & Kern D. Molecular Mechanism of Pin1-Tau Recognition and Catalysis. J. Mol. Biol. 2016
- PMID: 26996941
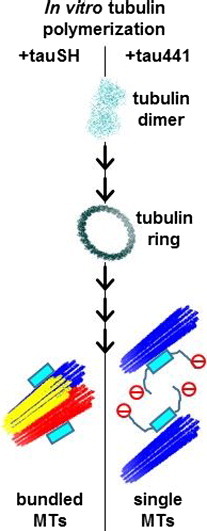
Kutter S, Eichner T, Deaconescu AM & Kern D. Regulation of Microtubule Assembly by Tau and not by Pin1. J. Mol. Biol. 2016
- PMID: 26996940
Pontiggia F, Pachov DV, Clarkson MW, Villali J, Hagan MF, Pande VS & Kern D. Free energy landscape of activation in a signaling protein at atomic resolution. Nat. Commun. 2015
Agafonov RV, Wilson C & Kern D. Evolution and intelligent design in drug development. Front. Mol. Biosci. 2015
- PMID: 26052517
- PMCID: PMC4440380
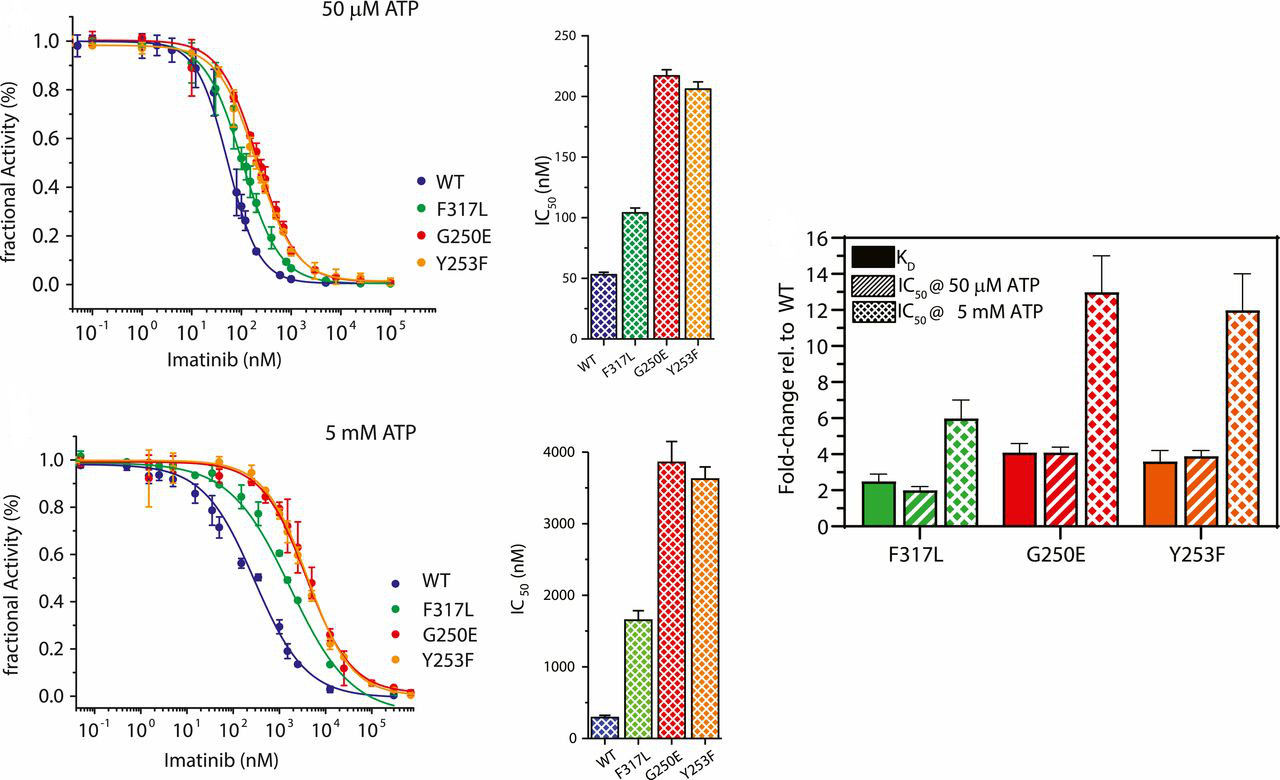
Wilson C#, Agafonov RV#, Hoemberger M, Kutter S, Zorba A, Halpin J, Buosi V, Otten R, Waterman D, Theobald DL & Kern D. Using ancient protein kinases to unravel a modern cancer drug’s mechanism. Science 2015
- PMID: 25700521
- PMCID: PMC4405104
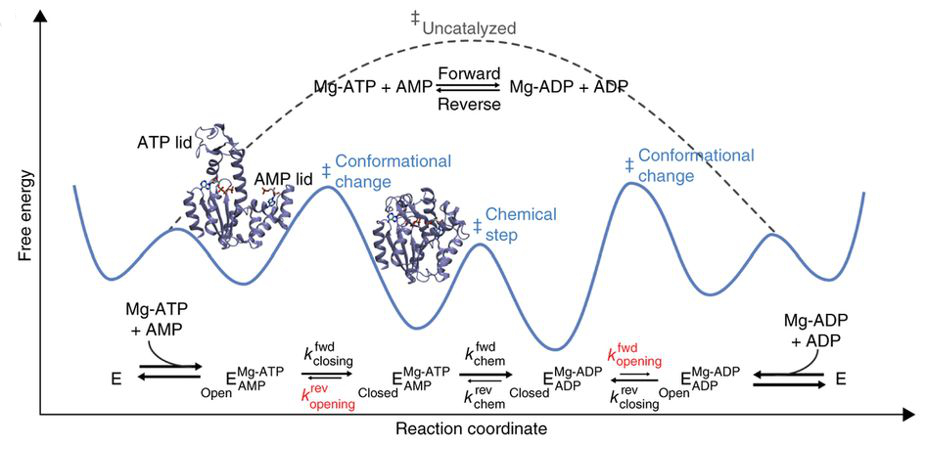
Kerns SJ#, Agafonov RV#, Cho YJ#, Pontiggia F, Otten R, Pachov DV, Kutter S, Phung LA, Murphy PN, Thai V, Alber T, Hagan MF & Kern D. The energy landscape of adenylate kinase during catalysis. Nat. Struct. Mol. Biol. 2015
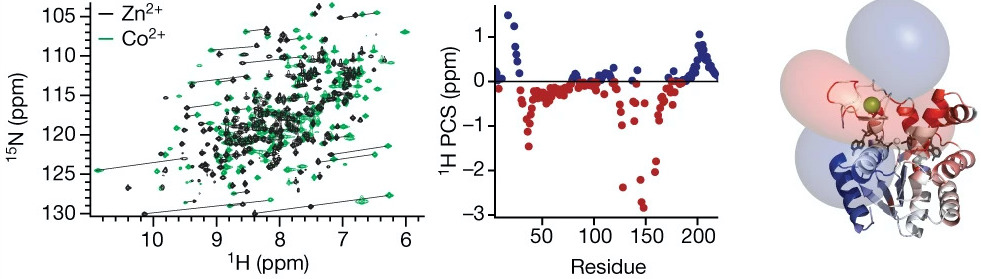
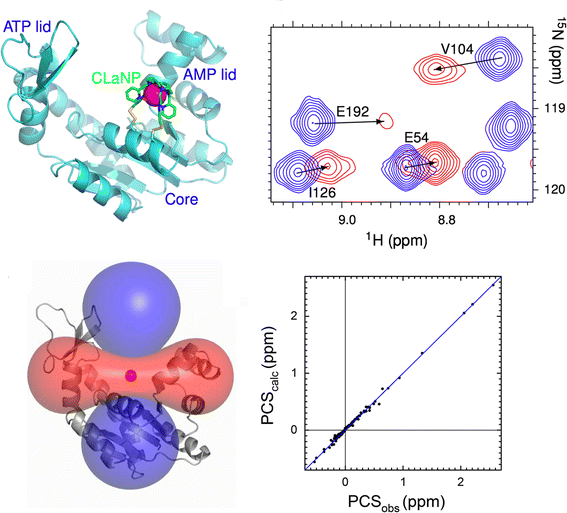
Hass MA, Liu WM, Agafonov R.V., Otten R, Phung LA, Schilder JT, Kern D & Ubbink M. A minor conformation of a lanthanide tag on adenylate kinase characterized by paramagnetic relaxation dispersion NMR spectroscopy. J. Biomol. NMR 2015
- PMID: 25563704
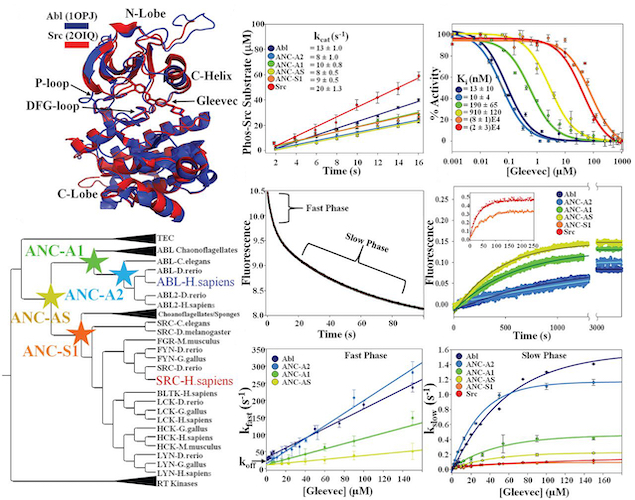
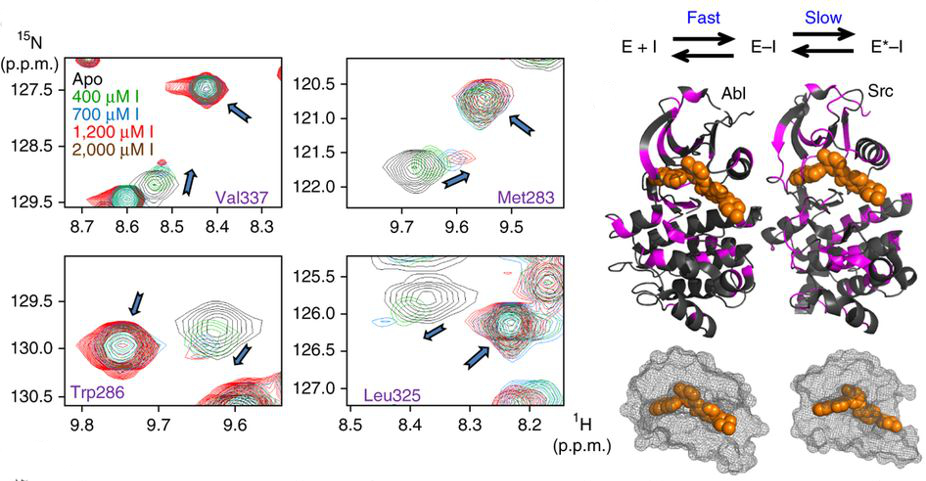
Agafonov RV#, Wilson C#, Otten R, Buosi V & Kern D. Energetic dissection of Gleevec’s selectivity toward human tyrosine kinases. Nat. Struct. Mol. Biol. 2014
- PMID: 25218445
- PMCID: PMC4266587
Zorba A#, Buosi V#, Kutter S, Kern N, Pontiggia F, Cho YJ & Kern D. Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife 2014
Villali J, Pontiggia F, Clarkson MW, Hagan MF & Kern D. Evidence against the “Y-T coupling” mechanism of activation in the response regulator NtrC. J. Mol. Biol. 2014
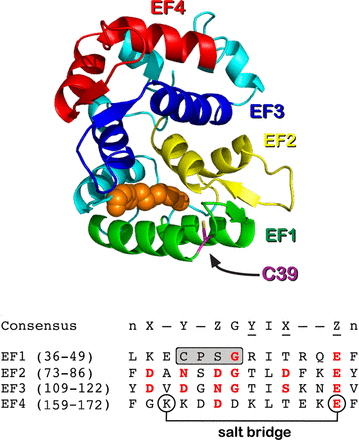
Ranaghan MJ, Kumar RP, Chakrabarti KS, Buosi V, Kern D & Oprian DD. A Highly Conserved Cysteine of Neuronal Calcium-sensing Proteins Controls Cooperative Binding of Ca2+ to Recoverin. J. Biol. Chem. 2013
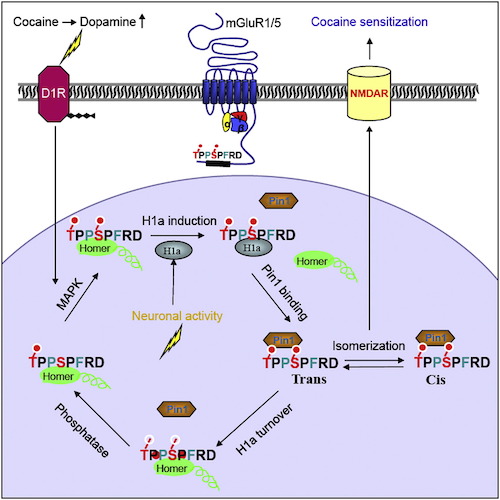
Park JM, Hu J-H, Milshteyn A, Zhang, P-W, Moore CG, Park S, Datko MC, Doming RD, Reyes CM, Wang XJ, Etzkorn FA, Xiao B, Szumlinski KK, Kern D, Linden DJ & Worley PF. A Prolyl-isomerase Mediates Dopamine-dependent Plasticity and Cocaine Motor Sensitization. Cell 2013
- PMID: 23911326
- PMCID: PMC3785238
Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T & Henzler-Wildman KA. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 2012
- PMID: 22178925
- PMCID: PMC3253143
Otten R, Villali J, Kern D & Mulder FAA. Probing Microsecond Time Scale Dynamics in Proteins by Methyl 1H Carr−Purcell−Meiboom−Gill Relaxation Dispersion NMR Measurements. Application to Activation of the Signaling Protein NtrCr. J. Am. Chem. Soc. 2010
- PMID: 21058670
- PMCID: PMC2991065
Villali J & Kern D. Choreographing an enzyme’s dance. Curr. Opin. Chem. Biol. 2010
- PMID: 20822946
- PMCID: PMC2987738
Gardino AK, Villali J, Kivenson A, Lei M, Liu CF, Steindel P, Eisenmesser EZ, Labeikovsky W, Wolf-Watz M, Clarkson MW & Kern D. Transient Non-native Hydrogen Bonds Promote Activation of a Signaling Protein. Cell 2009
- PMID: 20005804
- PMCID: PMC2891250
Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D & Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature 2009
Shim S, Yuan JP, Kim JY, Zeng W, Huang G, Milshteyn A, Kern D, Muallem S, Ming G-L & Worley PF. Peptidyl-Prolyl Isomerase FKBP52 Controls Chemotropic Guidance of Neuronal Growth Cones via Regulation of TRPC1 Channel Opening. Neuron 2009
- PMID: 19945390
- PMCID: PMC2786904
Clarkson MW, Lei M, Eisenmesser EZ, Labeikovsky W, Redfield A & Kern D. Mesodynamics in the SARS nucleocapsid measured by NMR field cycling. J. Biomol. NMR 2009
- PMID: 19641854
- PMCID: PMC2728245
Lei M, Velos J, Gardino A, Kivenson A, Karplus M & Kern D. Segmented Transition Pathway of the Signaling Protein Nitrogen Regulatory Protein C. J. Mol. Biol. 2009
- PMID: 19576227
- PMCID: PMC2765875
Thai V, Renesto P, Fowler CA, Brown DJ, Davis T, Gu W, Pollock DD, Kern D, Raoult D & Eisenmesser EZ. Structural, Biochemical, and in Vivo Characterization of the First Virally Encoded Cyclophilin from the Mimivirus. J. Mol. Biol. 2008
Henzler-Wildman KA & Kern D. Dynamic personalities of proteins. Nature 2007
- PMID: 18075575
Henzler-Wildman KA, Vu T, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hübner CG & Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature 2007
- PMID: 18026086
Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M & Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 2007
- PMID: 18026087
Gardino AK & Kern D. Functional Dynamics of Response Regulators Using NMR Relaxation Techniques. Methods Enzymol. 2007
- PMID: 17609130
Labeikovsky W, Eisenmesser EZ, Bosco DA & Kern D. Structure and Dynamics of Pin1 During Catalysis by NMR. J. Mol. Biol. 2007
- PMID: 17316687
- PMCID: PMC2975599
Wemmer DE & Kern D. Beryllofluoride Binding Mimics Phosphorylation of Aspartate in Response Regulators. J. Bacteriol. 2005
- PMID: 16321925
- PMCID: PMC1316999
Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE & Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature 2005
- PMID: 16267559
Tittmann K, Neef H, Golbik R, Hübner G & Kern D. Kinetic Control of Thiamin Diphosphate Activation in Enzymes Studied by Proton-Nitrogen Correlated NMR Spectroscopy. Biochemistry 2005
- PMID: 15952776
Kern D, Eisenmesser EZ & Wolf-Watz M. Enzyme Dynamics During Catalysis Measured by NMR Spectroscopy. Methods Enzymol. 2005
- PMID: 15808235
Wolf-Watz M, Thai V, Henzler-Wildman KA, Hadjipavlou G, Eisenmesser EZ & Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 2004
- PMID: 15334070
Bosco DA & Kern D. Catalysis and Binding of Cyclophilin A with Different HIV-1 Capsid Constructs. Biochemistry 2004
- PMID: 15147195
Kern D & Zuiderweg ERP. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003
- PMID: 14675554
Gardino AK, Volkman BF, Cho HS, Lee S-Y, Wemmer DE & Kern D. The NMR Solution Structure of BeF3--Activated Spo0F Reveals the Conformational Switch in a Phosphorelay System. J. Mol. Biol. 2003
Kern D. Cutting the leash. Nat. Struct. Biol. 2002
- PMID: 12080334
Bosco DA, Eisenmesser E, Pochapsky S, Sundquist WI & Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. USA 2002
- PMID: 11929983
- PMCID: PMC122755
Eisenmesser EZ, Bosco DA, Akke M & Kern D. Enzyme Dynamics During Catalysis. Science 2002
- PMID: 11859194
Kern G, Pelton J, Marqusee S & Kern D. Structural properties of the histidine-containing loop in HIV-1 Rnase H. Biophys. Chem. 2002
- PMID: 12034447
Volkman BF, Lipson D, Wemmer DE & Kern D. Two-State Allosteric Behavior in a Single-Domain Signaling Protein. Science 2001
- PMID: 11264542
Kern D, Volkman BF, Luginbühl P, Nohaile MJ, Kustu S & Wemmer DE. Structure of a transiently phosphorylated “switch” in bacterial signal transduction. Nature 1999
- PMID: 10622255
Kern D, Kern G, Neef H, Tittmann K, Killenberg-Jabs M, Wikner C, Schneider G & Hübner G. How Thiamin Diphosphate is Activated in Enzymes. Science 1997
- PMID: 8974393